BrainVoyager v23.0
DNN Tissue Classification
The DNN Segmentator can be launched from the DNN Segmentation dialog that can be invoked using the DNN Segmentation item in the Volumes menu.
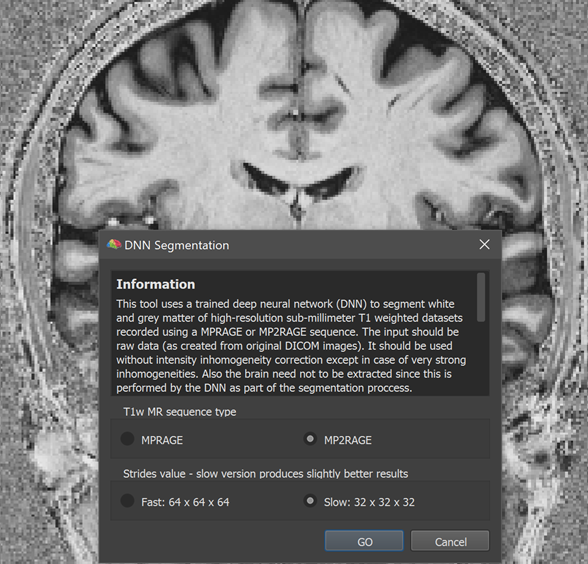
The screenshot above shows the lauched DNN Segmentation dialog on top of a high-quality 0.7 mm anatomical dataset recorded using the MP2RAGE image sequence (created from the original 'UNI' DICOM images without any subsequent processing). At the top of the dialog a text field presents information about the tool and the options available. The T1w MR sequence type field can be used to select one of the two currently supported imaging sequences, the standard MPRAGE sequence or the more advanced MP2RAGE sequence providing superior grey matter to white matter contrast. Since the dataset used here has been obtained with the MP2RAGE sequence, the corresponding option has been selected. The Strides value field can be used to select a version that produces results fast or slow. To explain the two options, it is relevant to know that the DNN operates on blocks (currently 64 x 64 x 64) of the full data and the Fast option visits each block only once. The Slow option (default) instead visits the same voxel multiple times by shifting the 64 block by only 32 voxels in each dimension. Visiting the same voxel multiple times leads to more robust results, especially when using data that has not been scanned with one of the trained sequences or not with 0.7 mm resolution.
Before launching the DNN Segmentation dialog, the program first checks whether the currently active document meets the following prerequisites:
- Spatial resolution . If the voxel size of the VMR does not have a resolution of 0.7 mm, the process may not provide good results. If the user wants to continue, the program will resample the data to 0.7 mm resolution before showing the dialog.
- 16-bit dataset . If there is not a V16 file with the same name in the same folder, DNN segmentation can not be performed. A V16 file contains the original data from the scanner when converting DICOM files to anatomical BV / NIfTI files without squeezing intensities into 256 (8-bit) values.
- MR pulse sequence . The segmentation DNN has been trained with MPRAGE and MP2RAGE MR sequences. While not checked at launch, one needs to select the appropriate sequence in the DNN Segmentation dialog. If other sequences have been used to obtain the anatomical data, results may not be useful.
Note that the anatomical volume should be available in BrainVoyager's standard (sagittal) orientation. In case the original scan is not recorded as a sequence of sagittal images, the (automatic) 'To Sagittal' transformation step should be performed when creating the dataset from DICOM files (or later); it is important, however, to not accept the suggestion of BrainVoyager to iso-voxel the data to 1 mm in order to keep the original (sub-millimeter) resolution of the dataset.
After making the choices about the MR sequence and speed-accuracy tradeoff, the segmentation process can be started by clicking the GO button. Note that in case the dimensions are not appropriate to run the DNN segmentation, the VMR / V16 volumes will be first reframed to supported dimensions. The filename of a reframed dataset can be recognized by the appended 'reframed' substring (e.g. [name]_reframed.vmr / .v16). The program then runs the deep neural network on the (reframed) input dataset using calls to TensorFlow from the embedded Python interpreter. The program indicates progress in the Python Console pane (see screenshot below).
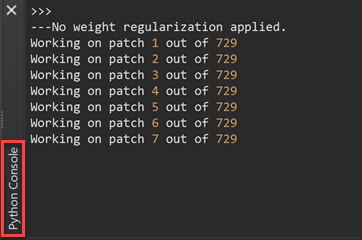
If you have specified the Slow option (recommended), there will be 600 - 800 steps required for calculating the tissue classification result while only about 100 - 150 steps are needed when selecting the Fast option (the exact number depends on the (reframed) dimensions of the data). Without an NVIDIA GPU, segmentation with the 'Fast' option wil take 5-10 minutes while the 'Slow' option requires about 30-60 minutes. With a GPU from NVIDIA (Windows, Linux) and appropriately prepared TensorFlow version, the Slow option takes only about 5 minutes on standard desktop computers.
When the DNN segmentation process has finished, results are presented as tissue probability volume maps that can be further processed in the automatically opened DNN Segmentation Postprocessing dialog. Postprocessing includes conversion of maps to non-overlapping volumes-of-interest (VOIs), disconnection of hemispheres, creation of standard segmentation volumes and reconstruction of cortex meshes.
Note. In some cases (especially when using datasets not recorded with a supported sequence and low resolution), some blocks of the data exhibit wrong labeling results, i.e. regions with grey and white matter might wrongly be classified as 'background'. If this is observed with the Fast option, make sure to rerun the segmentation with the Slow (highest accuracy) option to check whether this issue can be resolved.
Copyright © 2023 Rainer Goebel. All rights reserved.