BrainVoyager v23.0
MNI Normalization
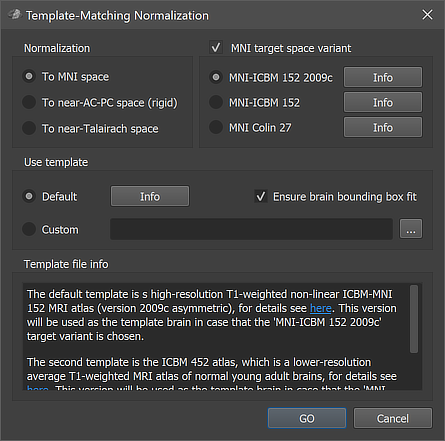
Anatomical 3D data sets can be normalized to MNI space using the Template-Matching Normalization dialog (see screenshot above) that can be invoked by clicking the Normalize To MNI Template Space item in the Volumes menu. The VMR document uses as input (i.e. the active document when calling the dialog) should have been preprocessed with the intensity inhomogeneity correction procedure including the (default) brain extraction step. A preprocessed VMR file can be usually identified by the "_IIHC" substring in its file name. Besides producing a VMR file in MNI (or a related) space, the normalization procedure also produces a spatial transformation (.TRF) file that can be used to transform (same-session) functional (.FMR) datasets into MNI space (see below). Anatomical and functional MNI space normalization can also be performed for all datasets of a project at once by specifying a corresponding normalization workflow using the data analysis manager or prgrammatically using the provided Python API.
Default and Custom Templates
There are unfortunately several variants of MNI space. As default, BrainVoyager uses the most widely used MNI-152 space represented by the high-resolution MNI-ICBM 152 2009c or lower resolution MNI-ICBM 152 target variant options in the MNI target space variant field. This space is recommended for volumetric whole-brain group analyses. Another possibility is the MNI Colin 27 option in this field which will normalize to a slightly different MNI space represented by the average of 27 anatomical scans of subject "Colin". Clicking the Info buttons will provide more information about a certain space including a link to the web sites with the original MNI/ICBM datasets. The input VMR will be aligned to the default template, which is either the mentioned 'MNI-ICBM 152 2009c' (if selected as the target variant), or the ICBM 452 average brain for all other variants. The input VMR is transformed to the MNI 152 2009c or the ICBM 452 template by mimimizing a cost function that reflect the match of the input VMR with the template VMR. From the ICBM 452 space, the final target space is reached by adding an additional pre-configured spatial transformation adjustment step to the obtained affine transformation matrix. Aligning to the ICBM 452 with subsequent addition of a transformation to the target space allows to create transformation files for multiple target spaces after a single template matching procedure; in previous versions, 3 transformation files were produced with names containing the original VMR base name ("[Input-VMR"]) and substrings to indicate the target spaces MNI-152 ("[Input-VMR]_TO_MNI_a12.trf"), MNI-Colin27 ("[Input-VMR]_TO_MNIColin27_a12.trf") and the base transformation to the default template brain ("[Input-VMR]_TO_ICBM452_a12.trf"). To avoid name clutter, only the .TRF file for the selected space is now produced.. Note that the "a12" substring in the .TRF names indicates that the transformation matrix has been obtained by fitting 12 parameters (3 rotations, 3 scales, 3 shears and 3 translations). One can use the stored .TRF files to transform the input VMR - or another VMR in the same-session native space - to one of the indicated target spaces without rerunning the normalization step. One of the output files, usually the "[Input-VMR]_TO_MNI_a12.trf" file (or another variant, see below), is also needed for transforming functional .FMR data into MNI space (see below).
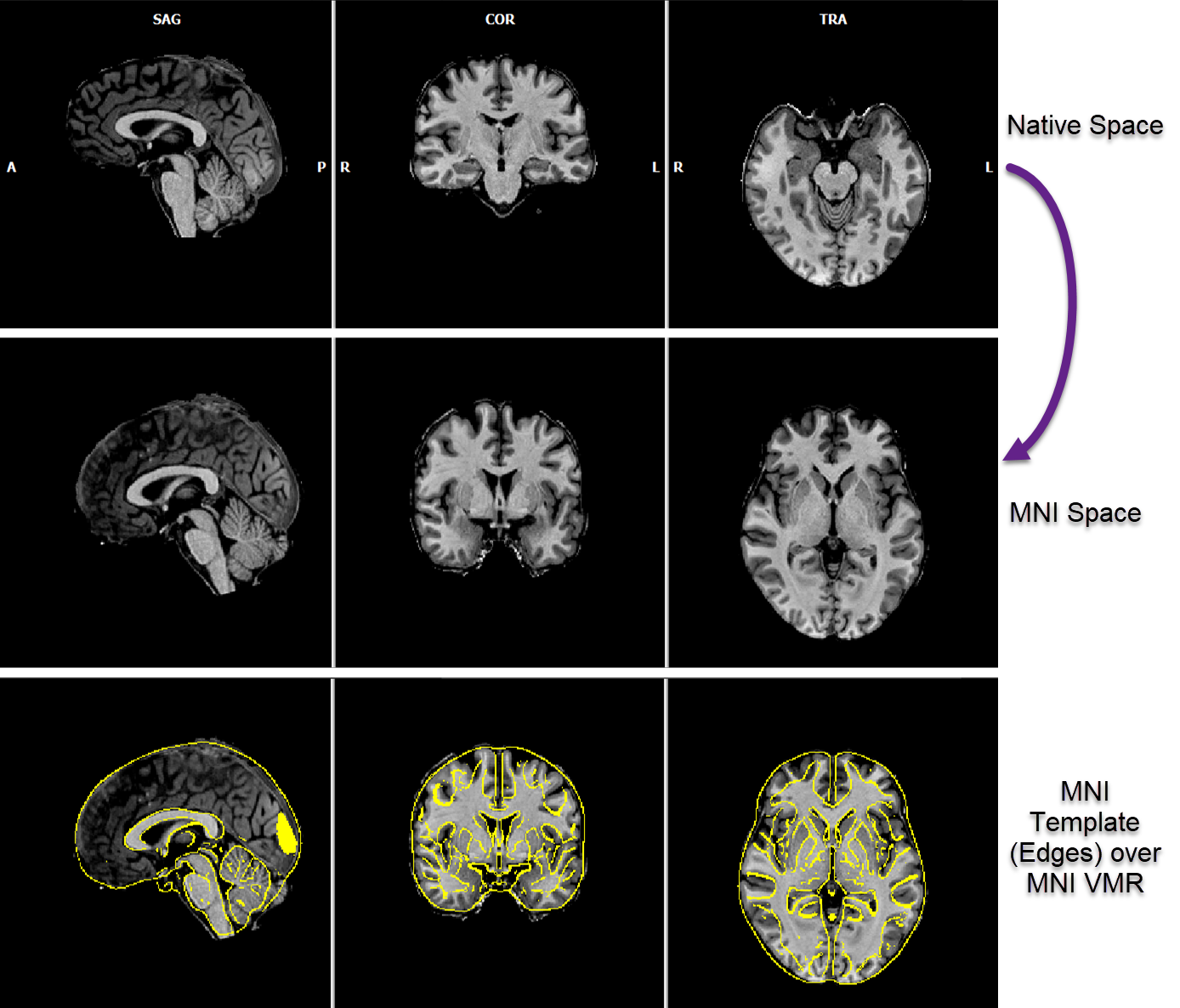
When inspecting coordinates in a MNI normalized VMR data set, the displayed coordinates are "MNI coordinates" with an origin (point with a zero value for the x, y and z coordinates, see snapshot below) that is close but not exactly located at the AC point as in the case of landmark-based Talairach space.
Since BrainVoyager 22.2, it is possible to select also other brains as the target for template matching by selecting the Custom option in the Use template field. Using, for example, the Colin template brain as a custom target brain will directly align a source brain to the Colin brain. With the custom template option It is possible to use a specific template brain as a better suited target for special applications, for example, when analyzing data from a children population. When using a custom target, the output VMR file can be identified by the "_cMNI.vmr" substring (instead of "MNI.vmr" for the two default templates) and the .TRF file can be identified by the "_TO_cMNI_a12.trf" substring. In case of an extra bounding box adjustment step (see below), the output TRF files will also contain a trailing "_adjBBX" substring. Note that in case that a V16 file is found with the same base name as the input VMR file, the (default or custom) normalization is also applied to this data set producing a (c)MNI .v16 output file.
Display for Quality Assurance
Template-matching of an input brain (usually in native space, see row 1 in the figure above) with a default (or custom) target brain is started when clicking the GO button in the dialog. Information about the progress of the procedure is reported in the Log pane. After a few seconds, the resulting MNI representation of the input brain is shown in the BrainVoyager multi-document area (see second row in the figure above). In order to allow verification of the normalization result, a blend of the contours of the template brain with the VMR document is shown automatically. For this display, the program loads a version of the respective template brain in MNI-152 space (from the 'Documents/BrainVoyager/MNITemplates' folder) that highlights its edges. This edge version is drawn in yellow over the normalized individual brain and makes it possible to see the normalized VMR document in relation to the contours of the template brain (see bottom row in the figure above). One can switch, as usual, between the two VMR documents and the blended view by using the standard VMR keyboard toggling shortcuts (F8, F9) or the respective Show primary VMR / Show secondary VMR / Blend 2 options in the Spatial Transf tab of the 3D Volume Tools dialog.
Initial Rotation to Near-ACPC and Talairach Space
In BrainVoyager 22.2, a preparatory rigid rotation step has been added to the template matching procedure to improve the normalization result for input datasets that have been scanned in an orientation that deviates largely with respect to the axial near AC-PC plane of the template brain. The figure below (first row) shows an example of such a case, which was not optimally aligned in previous versions. The preparatory step performs an iterative initial rigid (6 parameter) alignment to bring a source brain close to the AC-PC plane orientation of the template brain. The procedure then continues with a 9 and finally (12 parameter) affine alignment as in previous versions. The initial rigid "near AC-PC" files are temporary stored to disk but deleted when continuing with the non-rigid transformation. It is, however, possible to obtain the brain in this near AC-PC space as output by turning on the To near-AC-PC space (rigid) option in the Normalization field of the Template-Matching Normalization dialog. The output file can be identified by the "_nACPC" suffix of the produced VMR and spatial transformation (TRF) file. This output may be useful as a robust automatic alignment to (near) AC-PC space for applications such as neuronavigation that do not want to change the shape and size of the brain. In case one wants to transform a brain into "full" AC-PC or Talairach space, the rigid template matching to nACPC space can also be used as a preparatory step for the subsequent application of the precise (automatic or manual) landmark-based AC-PC and Talairach transformation procedure.
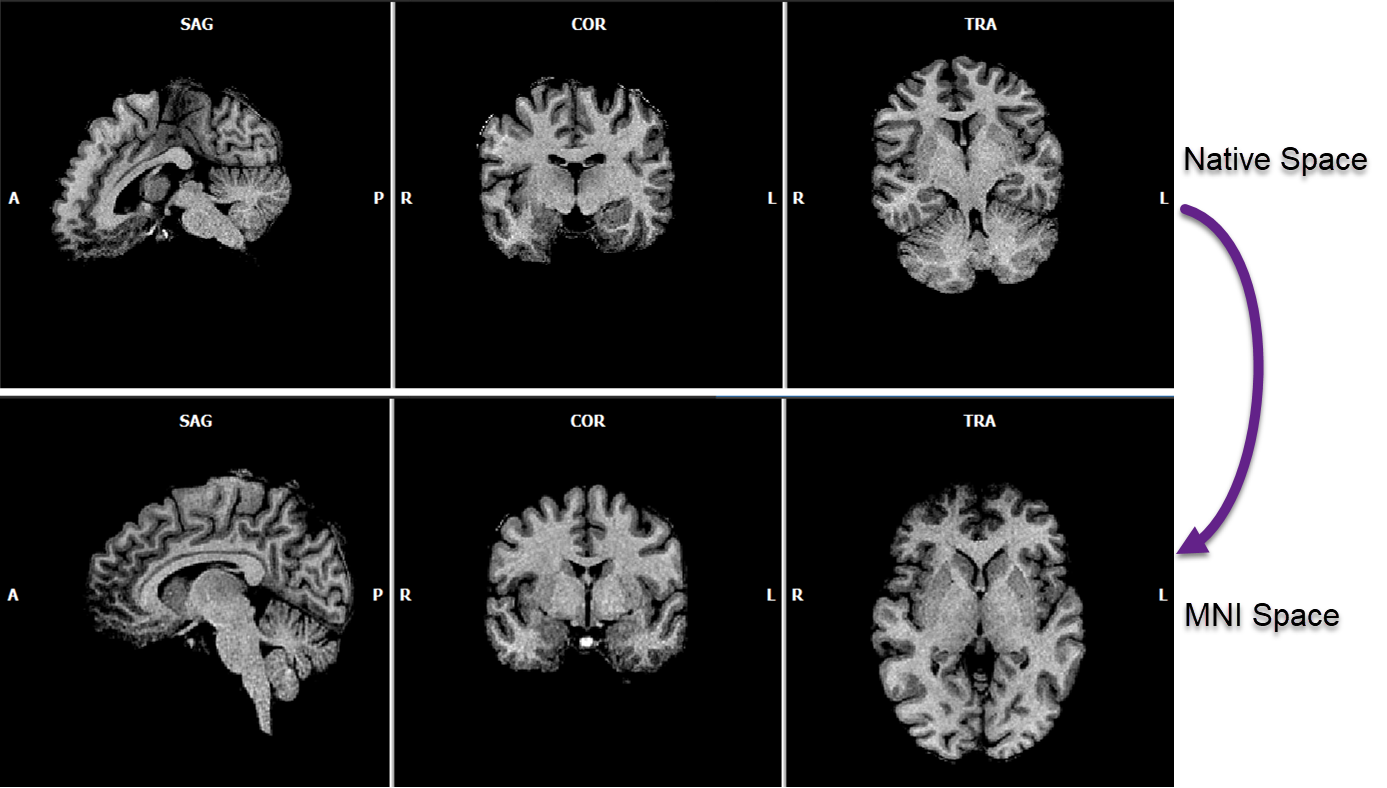
Template matching with the ICBM 452 can also be used to transform an input VMR data set to "near Talairach" space when selecting the To Talairach Space option. In this case the ICBM 452 template is used for template matching but then a fixed ACPC and TAL transformation step are appended to bring the data into near Talairach space. After running template matching, the two files "_mACPC.trf" and "_mACPC.tal" are stored to disk that can be used to transform functional data into Talairach space when using the Create VTC dialog (see below). Since the resulting transformation files are not obtained with the classical landmark-based ACPC-TAL procedure, the transformation files and the resulting VMR file will be marked by adding letter "m" (e.g. "_mTAL.vmr") to indicate that this Talairach transformation is the result of a template matching procedure. The resulting VMR file will usually be in a space that is close, but not identical. to a VMR file that has been normalized using the conventional Talairach procedure; most notably, the distance between the AC and PC point will usually not correspond to the standardized distance of a conventional landmark-based procedure.
Non-Linear Bounding-Box Adjustment
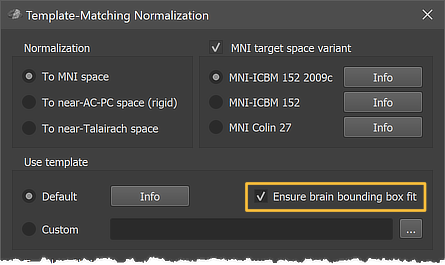
Since BrainVoyager 22.2 it is possible to perform an extra bounding box adjustment step that is performed after the 12 parameter affine transformation (see marked Ensure brain bounding box fit option in the figure above). This step (used as default) has been introduced to ensure that brains fit better into the standard (or custom) MNI bounding box. Without this step, it has been observed in some cases that the borders of a transformed brain did not exactly match the borders of the target brain's bounding box, i.e. the brain either extended a bit beyond or did not reach the bounding box. To ensure matching the MNI bounding box, a global non-linear adjustment is performed that scales 6 sub-cuboids of the transformed brain into the MNI bounding box. This is a similar piecewise linear (globally non-linear) procedure as used when going from AC-PC space to Talairach space except that the AC-PC distance is ignored in the MNI case. The sub-cuboids are defined by measuring the distance from the center (near AC) point to the left / right, upper / lower and front /back border of the transformed brains. From these 6 distance values 6 scaling values are computed by relating the measured distances to the default distances of the MNI (or custom template) bounding box.
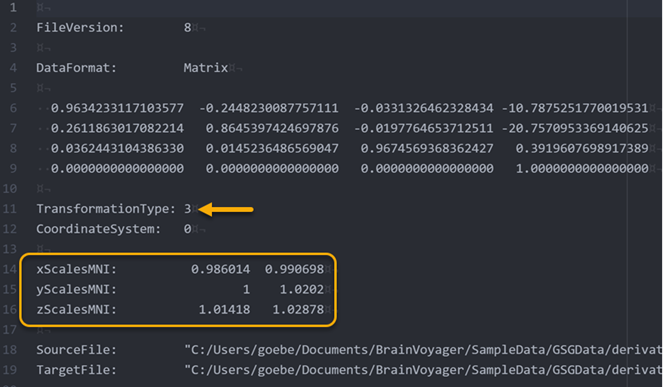 The 6 calculated scaling values are also stored in the produced spatial transformation TRF file (see rectangle in figure above) so that the scaling operation will also be applied when bringing functional data into MNI space. The value '3' for the 'TransformationType' entry (see arrow in the figure above) indicates that the stored transformation includes the combination of a 12-parameter affine matrix (see 4x4 values below 'DataFormat: Matrix') and a subsequent piecewise linear scaling operation determined by the provided 6 scaline values. To identify whether a transformation file contains scaling parameters, the produced file name contains the "_adjBBX" substing, i.e. the file name will be "[Input-VMR]_TO_MNI_a12_adjBBX.trf" instead of "[Input-VMR]_TO_MNI_a12.trf".
The 6 calculated scaling values are also stored in the produced spatial transformation TRF file (see rectangle in figure above) so that the scaling operation will also be applied when bringing functional data into MNI space. The value '3' for the 'TransformationType' entry (see arrow in the figure above) indicates that the stored transformation includes the combination of a 12-parameter affine matrix (see 4x4 values below 'DataFormat: Matrix') and a subsequent piecewise linear scaling operation determined by the provided 6 scaline values. To identify whether a transformation file contains scaling parameters, the produced file name contains the "_adjBBX" substing, i.e. the file name will be "[Input-VMR]_TO_MNI_a12_adjBBX.trf" instead of "[Input-VMR]_TO_MNI_a12.trf".
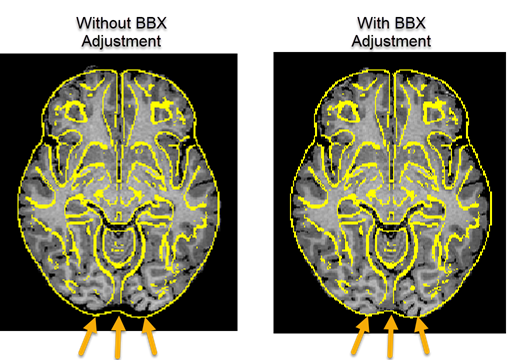
While the scaling values are usually very close to 1.0, the figure above shows an example of the effect of bounding box adjustment in a more extreme case. The arrows on the left indicate that the brain does not extend to the posterior end of the brain when performing MNI template matching without the extra adjustment step, while the brain is well aligned to the posterior boundary of the template brain when including the bounding box adjustment step (right side in figure above). In combination with the initial rigid rotation step, the updated MNI normalization routine produces robust results.
MNI Normalization of FMR Data
Functional .FMR datasets can be transformed into MNI space using the standard Create VTC dialog that can be invoked by clicking the Create Normalized VTC From FMR Data item in the Analysis menu. Furthermore, MNI-space VTCs can also be created using the FMR normalization workflow in the context of the data analysis management tools.
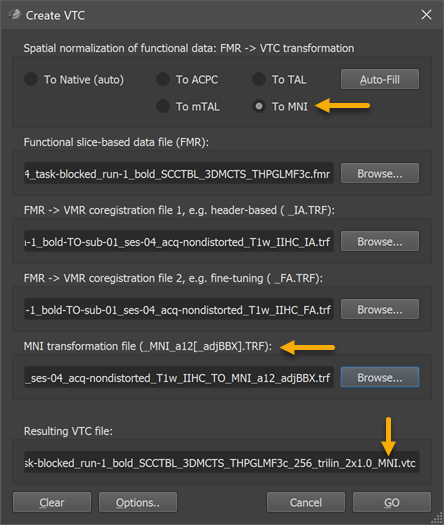
As with transformation of FMR data sets to other spaces, the input-FMR data set need to be specified in the Functional slice-based data file (FMR) field using the respective Browse button. Also the the FMR-VMR initial and fine-tuning alignment .TRF files need to provided in the FMR - VMR coregistration file 1 and FMR - VMR coregistration file 2 fields as usual. In order to prepare the dialog for MNI normalization, the To MNI option need to be selected (see arrow at top n snapshot above). This will adjust the dialog showing the MNI tranformation file (_MNI_a12[_adjBBX].TRF) field (see arrow in the middle of the snapshot above); the TRF file stored as a result of MNI normalization of the hosting VMR data set needs to be specified here, usually the .TRF file with the name "[Input-VMR]_TO_MNI_a12_adjBBX.trf" or "[Input-VMR]_TO_MNI_a12.trf" in case that the bounding box adjustment step is not used. If other target spaces are used, the respective .trf file need to be specified in the same way. The FMR-VMR alignment files will bring the functional data in the space of the hosting VMR, which should be a 1mm space. The 12-parameter transformation matrix (and 6 bounding box scaling values) contained in the MNI normalization file will further transform the data from the space of the hosting VMR to MNI space in the same way as has been done before with the anatomical dataset. Internally all transformation matrices are concatenated and the transformation from the original FMR space to MNI space is performed in a single transformation step for each functional volume of the FMR data. The name of the output VTC file is shown in the Resultig VTC file field; note that the suggested name for the VTC data set contains the substring "_MNI" (see arrow in snapshot above) indicating that the resulting data will be normalized in MNI space.
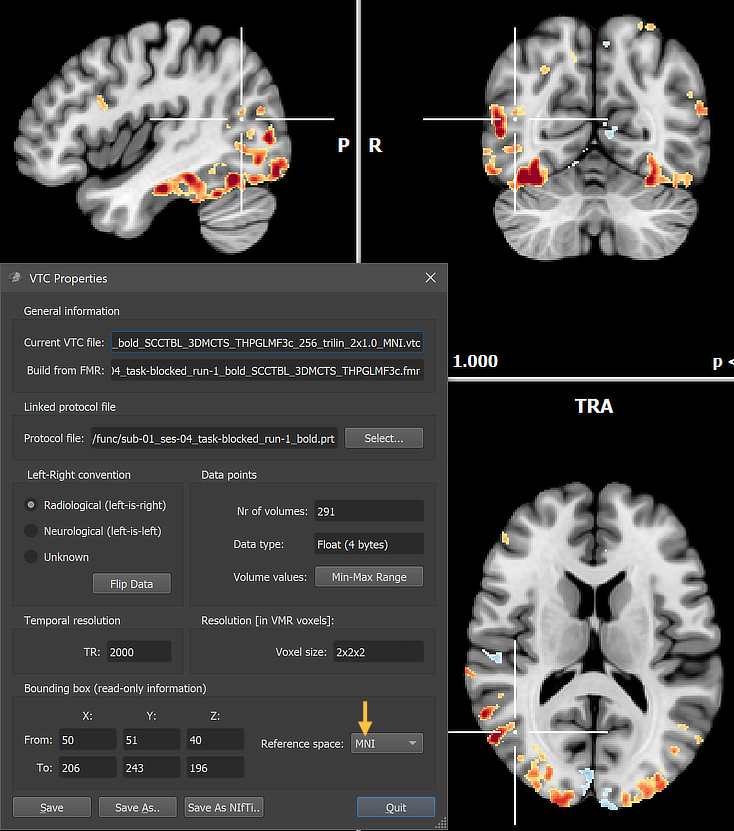
The resulting VTC file can be linked to an individual VMR (e.g. "[Input-VMR]_MNI.vmr" or an average (template) VMR data set in MNI space. In the example snapshot above, the created MNI space VTC has been linked to the high-resoluton template brain "MNI_ICBM152_T1_NLIN_ASYM_09c_BRAIN.vmr" that is available in the "MNITemplates" sub-folder in the user's "Documents/BrainVoyager" directory and can be easily loaded by using the Open MNI VMR item of the File menu. The snapshot also shows that the VTC Properties dialog (like the VMR Properties dialog) can be inspected to verify that the functional VTC data is in MNI space (see arrow in figure above).
Transforming High-Resolution Functional Data to MNI Space
Transforming high-resolution (sub-millimeter) FMR-STC data to 1 mm MNI space is possible since BV 22.4.2 even when using a non-1mm (e.g., 0.7mm) anatomical VMR for the FMR-VMR coregistration step. For the anatomical MNI transformation, one must, however, create (using the iso-voxelation dialog) and use a 1mm VMR as described above. For VTC creation in MNI space, one needs to use the 1mm VMR as the hosting (loaded) VMR. One then provides the IA and FA TRF files that were created using the (non-1mm) FMR-VMR coregistration step and the MNI file used for 1mm MNI transformation. The program then ensures that the correct original VMR voxel size (e.g, 0.7 mm) is used for the IA-FA coregistration step of the functional data despite the fact that the loaded VMR has a different (1 mm) resolution. This is possible because the IA TRF file stores the original voxel resolution which was used for FMR-VMR coregistration (while rotations are independent of voxel size, the scaling to the original VMR resolution for coregistration ensures that translations are performed in the correct resolution). The result of coregistration is then scaled to the 1mm space of the hosting VMR followed by the MNI transformation step. As described above, all these steps are concatenated into a single affine transformation matrix. As an extra step, It is also planned ifor version 23 of BrainVoyager to add the possibility to scale the MNI space to a higher resolution (e.g. 0.7). This will allow statistical group VTC analysis in non-1mm MNI space if done consistently across subjects.
Copyright © 2023 Rainer Goebel. All rights reserved.